In a new report about the possible origin of SARS-CoV-2 an independent investigator from the DRASTIC group challenges claims that Covid-19 emerged from the Huanan market in Wuhan via zoonosis and argues robustly that SARS-CoV-2 had a laboratory origin.
“The experimental practices at WIV [the Wuhan Institute of Virology], when considered alongside genetic and historical evidence, strongly support a lab-leak origin for SARS-CoV-2,” the report states.
“This challenges the dominant zoonotic hypothesis and underscores the urgent need for renewed scrutiny of biosafety protocols within virology research.”
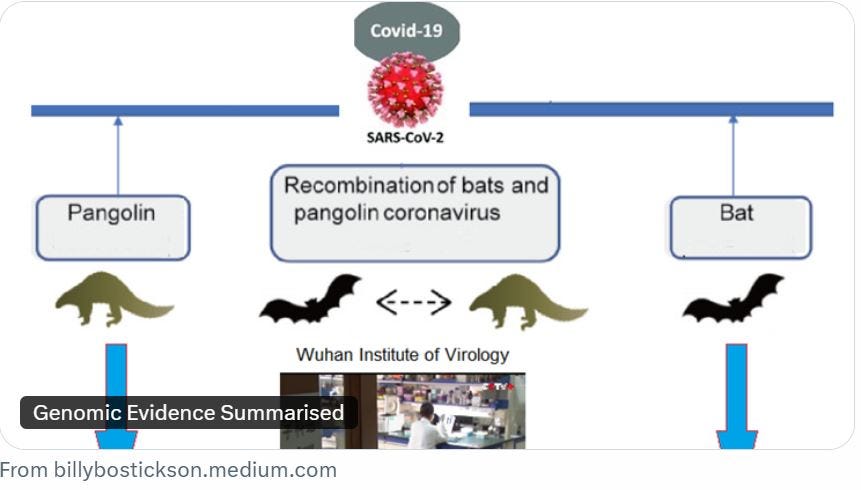
In the 206-page report, entitled ‘The Pangolin (Coronavirus) Papers’, the author, who writes and posts under the name Billy Bostickson, states: “This report hypothesises that the activities at WIV, involving Guangdong (GD) Pangolin-CoVs, RaTG13, and other unpublished bat coronaviruses in cell cultures, culminated in recombination and horizontal gene transfer, leading to the emergence of SARS-CoV-2.”
Bostickson says that new data reveal that the WIV possessed pangolin coronaviruses like GD/2019, with near-identical receptor-binding domains (RBDs) to SARS-CoV-2, by 2019, alongside bat viruses with FCS-like insertions, “suggesting a lab-mediated recombination event”.
He proposes that SARS-CoV-2 acquired an FCS (furin cleavage site), which has been flagged as a potential indicator of genetic manipulation, through recombination with a pangolin coronavirus at the WIV in 2019.
“Such an event, facilitated by cell culture contamination and experimental animals, may have resulted in an unintended laboratory release ...," he writes
Bostickson notes that the FCS enhances the virus’s capacity to infect human cells.
"The absence of a furin cleavage site in natural pangolin coronaviruses, juxtaposed with the Wuhan Institute of Virology’s extensive research, tilts the scales toward a laboratory origin for SARS-CoV-2, a conclusion not easily brushed aside," he writes.
He cites Zhang et al. (2020), who observed that only SARS-CoV-2 possesses a furin cleavage site at the S1/S2 boundary, which, he says, suggests a unique mechanism for host cell entry possibly derived from recombination with other coronaviruses.
“They provided compelling evidence supporting the hypothesis of SARS-CoV-2 emerging from a recombination between a pangolin coronavirus and a bat coronavirus in cell lines in 2018, with a lab leak in autumn 2019...,” Bostickson writes.
It is now clear, he says, that counter arguments from those who say SARS-CoV-2 had a zoonotic origin have failed, “with no pangolin trade at Huanan (Xiao et al., 2020), the absence of outbreaks along smuggling routes (Lancet, 2020) and no intermediate host identified after five long years of searching ...".
Bostickson cites “negative SARS-CoV-2 tests in animals, the absence of precursor sequences, the dominance of lineage B, and evidence of viral circulation by September to October 2019, well before the market cases were reported”.
All this, he says, suggests that the market was a superspreader site rather than the place where Covid-19 originated.
Bostickson notes that communications from 2020, unearthed by Francisco de Asis and other members of the DRASTIC team, indicate that prominent scientists initially entertained the plausibility of a lab leak.
“They noted WIV's passaging of pangolin CoVs and RaTG13, but political pressures from figures such as Anthony Fauci and Francis Collins later shifted the narrative towards a natural origin,” Bostickson writes.
Genetic analysis shows that the RBD of GD Pangolin-CoV is nearly identical to that of SARS-CoV-2, necessitating minimal mutations for adaptation to humans, he adds.
“Mid-2018 recombination events with bat covs further facilitated the introduction of the FCS, enhancing human infectivity,” he writes.
The likely work at the WIV on RaTG13 and FCS insertions, as outlined in the DEFUSE project, alongside intelligence assessments from various agencies, suggests that a lab leak is the more plausible explanation for the origin of SARS-CoV-2, Bostickson says.
The ‘Defusing the Threat of Bat-borne Coronaviruses’ (DEFUSE) grant proposal was submitted to the Defense Advanced Research Projects Agency (DARPA) in the US by the EcoHealth Alliance (EHA) in March 2018.
The proposal, which was rejected, was made to DARPA under the umbrella of the PREventing EMerging Pathogenic Threats (PREEMPT) programme.
The EHA proposed injecting chimeric bat coronaviruses collected by researchers at the WIV into “batified” mice and humanised mice genetically altered to express the human ACE-2 receptor.
“Batified” mice are mice that have been irradiated and injected with bat bone marrow.
The DEFUSE proposal, which some scientists have described as a blueprint for SARS-CoV-2, included discussion about the planned introduction of human-specific cleavage sites into bat coronaviruses.
Bostickson cites claims that China’s People’s Liberation Army (PLA) has been driving bio-research at the WIV.
“Independent research over the past five years aligns with claims of a PLA-driven bio-research network at WIV that exploited pangolin CoVs (2017-2019) under military units like 63919 and AMMS, suggesting a militarised origin for SARS-CoV-2,” he writes.
“The laboratory-adapted traits of GX_WIV [a pangolin CoV-related strain in WIV datasets] and its near-identical sequence to GX_P2V [a lab-adapted pangolin CoV from Guangxi samples] provide compelling evidence that the Wuhan Institute of Virology conducted experiments with pangolin viruses, supporting a laboratory escape scenario.
“This report posits that the 99.78% sequence identity between GX_WIV and GX_P2V, along with its laboratory-adapted characteristics, reinforces the assertion that the Wuhan Institute of Virology engineered a precursor to SARS-CoV-2.”
Bostickson adds that GX_WIV exhibits mutations (e.g., K217N, D218N) indicative of cultivation in HEK293T cells, pointing to lab refinement rather than natural evolution.
He says that the WIV’s possession of unpublished bat CoV sequences, like those with FCS-like insertions (RmYN02), suggests there was a pool of genetic material available for recombination.
He cites a 2024 analysis by Samson et al., which reveals that a horizontal gene transfer of the spike gene (gene S) from bat coronaviruses (BANAL species) to pangolin coronaviruses occurred around mid-2018, “as evidenced by the phylogenetic proximity of these sequences to SARS-CoV-2 in the gene S and receptor-binding domain (RBD) phylogenies”.
He adds: “This recombination event accounts for the genetic similarity between SARS-CoV-2 and pangolin coronaviruses in the RBD region, a critical factor in the virus's ability to infect humans.”
Moreover, Bostickson writes, the timeline supports the hypothesis of a potential leak from the WIV, which held pangolin viral isolates in 2018–2019, possibly contributing to SARS-CoV-2’s evolution.
“The work at WIV with RaTG13, delayed sequence uploads, and FCS insertion plans in the DEFUSE proposal point towards a lab-mediated origin,” Bostickson concludes.
He adds that a 49-nucleotide sequence in Guangzhou pangolin CoV reads, containing a 12-nucleotide segment identical to SARS-CoV-2’s FCS, was identified in WIV datasets.
“This sequence’s atypical codon usage suggests synthetic manipulation rather than natural contamination,” Bostickson writes.
Inaccessible WIV databases and delayed sequence uploads, e.g., of RaTG13, indicate potential cover-ups, undermining transparency, he says.
“Reports persist about WIV withholding data, particularly pangolin coronavirus samples from that year, which heighten suspicion.”
This article is also available on my Changing Times website.
Six Changing Times articles about the possible lab origin of SARS-CoV-2 are cited in Billy Bostickson’s report.
All my articles are freely accessible, but I do need the support of my readers. If you like my articles, please do share them. If you wish to support my work financially with a donation please click this PayPal link.
If you would like to take out a paid subscription to my Changing Times website, there are PayPal subscription buttons on the website.
Thanks to all those who already support me and thanks in advance to those who will be supporting me in the future.